March 03, 2021
Tag:
Summary
Human amyloid has been shown to interact with viruses and interfere with virus replication. Based on this observation, we adopted a synthetic biology method to design virus-specific amyloid to combat influenza A virus and Zika virus proteins. Each amyloid protein shares a homologous and easily aggregated fragment with a specific viral target protein. For influenza, we demonstrated that anti-PB2 designed amyloids accumulate in influenza A infected tissues. In addition, this amyloid specifically targets influenza A virus and its common PB2 polymorphism, rather than influenza B virus lacking homologous fragments. Our amyloid model shows that the sequence specificity of amyloid interactions has the ability to adjust amyloid-virus interactions while allowing the flexibility to maintain activity in evolutionary variation.

Introduction
For decades, it has been mainly to study human starch as the main feature of disease protein deposition, but in recent years, functional starch has been found in several human tissues1 and the lives of all kingdoms, resulting in a revised view of the structural nature of amyloid. The above is toxic or sick. Recently, more and more evidence supports the hypothesis that disease-related amyloid may not only be a by-product of disease2, but they may actually have functional benefits, such as acting as human pathogen interactions , Including viruses. For example, studies have shown that the peptide associated with Alzheimer's disease (AD), amyloid (A), may play a role in innate immunity against herpes virus brain infection. More specifically, Eimer et al. showed that a molecule interacts with herpes glycoprotein to induce the formation of an amyloid. The dissemination of "micro-aggregation" leads to the trapping of virus particles, resulting in amyloid-driven antiviral effects. Whether the amyloid deposit formed by encountering the virus plays a role in the occurrence of AD remains to be determined, but herpes infection is a known risk factor for AD.
Polymerization catalysis, as observed when A[beta] binds to herpes viruses, well describes the process driven by so-called polymerization prone regions (APRs). These APRs participate in the same kind of interactions, forming tightly packed intermolecular amyloid structures, leading to a highly sequence-specific process, but allowing protein interactions to share highly homologous APRs. Interestingly, a coronary cell also shares a homologous APR sequence with envelope glycoprotein B. A herpes protein that has been shown to interact with an immune oligonucleotide. In order to study whether amyloid, which is homologous to a certain viral protein, can actually interact with the protein specifically to interfere with viral replication, we adopted synthetic biology methods. We designed two synthetic amyloid proteins that encode virus-specific APRs identified in influenza A and Zika virus (ZIKV) proteins. We found that every amyloid protein interferes with the replication of the corresponding virus without cross-reaction. For influenza A virus, we demonstrated that our synthesized amyloid aggregates at the site of infection and interferes with influenza A virus replication in a mouse infection model. Amyloid binds to a specific viral target protein in an APR-specific manner, forcing the protein to form a non-functional conformation. Influenza B viruses lacking APR homologues are not affected by synthetic amyloid, which highlights the sequence specificity of this interaction. We jointly proposed a design of fully synthetic amyloid, based only on the APR identified in the viral protein, showing specific antiviral properties. In addition, our results indicate that APR-mediated amyloid interactions have the same characteristics that define true functional biological interactions, including specificity and selectivity in vivo.
Conclusion: Design of a synthetic peptide with antiviral properties based on apr
In order to study the possibility of interaction between amyloid and viruses encoding homologous APR, we used a synthetic biology method, using influenza A virus as a model virus. In order to design a synthetic amyloid with homologous APR to influenza a protein, we first used the statistical thermodynamic algorithm TANGO to identify all APRs in the influenza a protein group. (Design of synthetic peptide with antiviral properties based on apr, see figure)
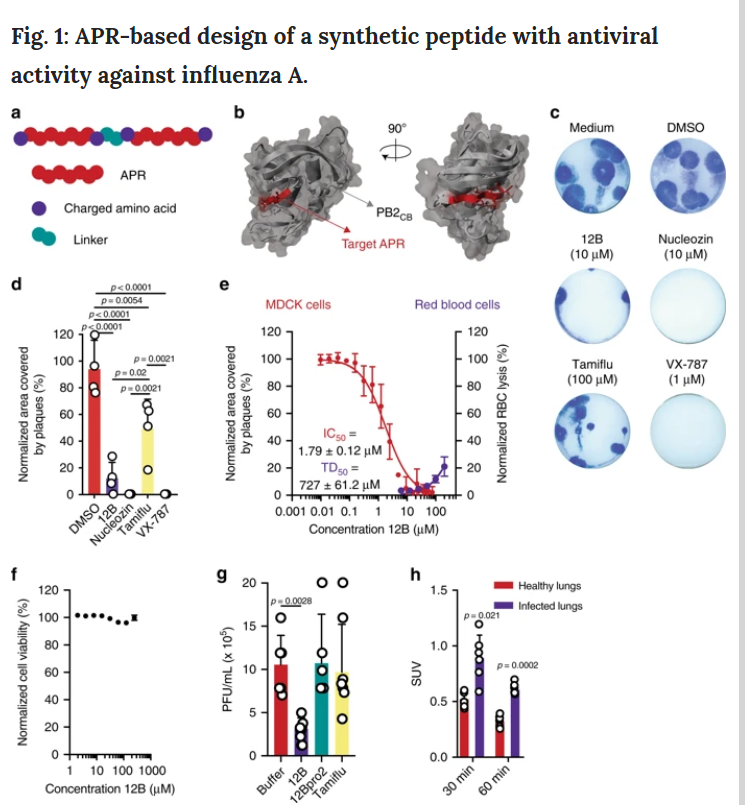
According to the design, the best scoring peptide sequence is WDLIQLIVSDGSDLIQLIVSD, which is called Peptide 12B, in which the target protein APR (381LIQLIVS387) has negative amino acid residues on both sides, and the n-terminal tryptophan is added for accurate determination. concentration.
Finally, we used two other experiments to verify the antiviral effect of peptide 12B: a multi-cycle replication experiment and protection against cytopathic effects (Supplementary Figure 1g, h). These two experiments have confirmed one point, Peptide 12B has antiviral effect on influenza A virus.
Explore the chemical space of antiviral synthetic starch
In order to further prove whether our method of treating influenza A can be transferred to other applications, we have set up to identify amyloid with ZIKV as the target. Using the same method of designing peptides as described above, the results showed that peptide R50 (RVAIAWLLRGSRVAIAWLLR), derived from the APR identified in membrane glycoprotein, showed the most significant effect.
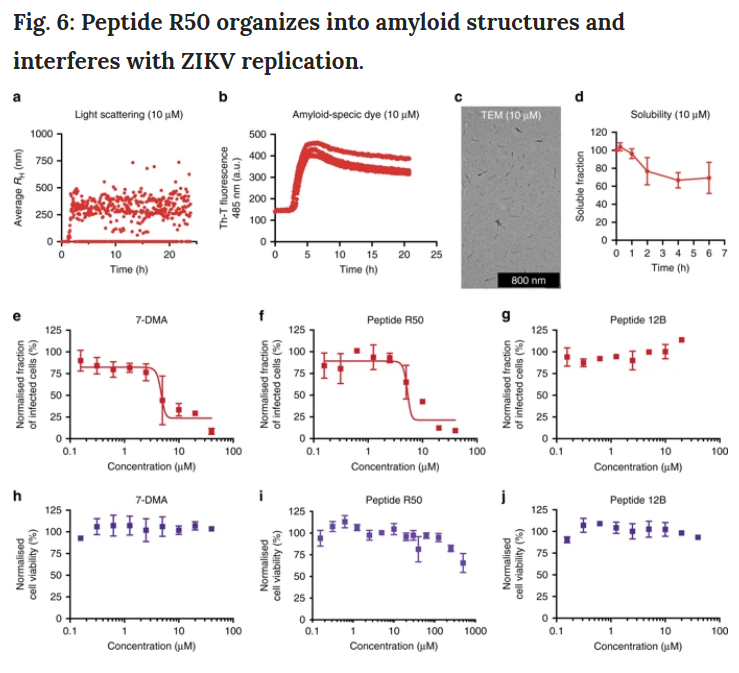
From these data, our design method is universally applicable and can be used to specifically target different viruses. In other words, these antiviral peptide families constitute a new chemical space, which can be used to mine new antiviral drugs and play their roles through targeted protein aggregation mechanisms.
Our company( www.gtpeptide.com ) is providing the related Conopeptide for research purpose, welcome to contact: sales@gotopbio.com for any requests.


Contact Us
Tel: (+86) 400 610 1188
WhatsApp/Telegram/Wechat: +86 13621645194
+86 15021993094
Follow Us:




 Pharma Sources Insight July 2025
Pharma Sources Insight July 2025


